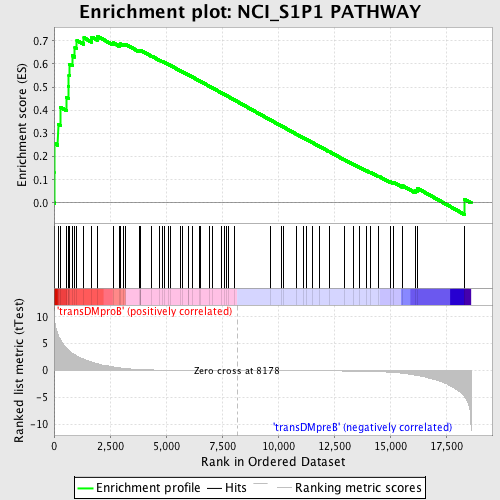
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_transDMpreB.phenotype_transDMproB_versus_transDMpreB.cls #transDMproB_versus_transDMpreB.phenotype_transDMproB_versus_transDMpreB.cls #transDMproB_versus_transDMpreB_repos |
| Phenotype | phenotype_transDMproB_versus_transDMpreB.cls#transDMproB_versus_transDMpreB_repos |
| Upregulated in class | transDMproB |
| GeneSet | NCI_S1P1 PATHWAY |
| Enrichment Score (ES) | 0.7201808 |
| Normalized Enrichment Score (NES) | 1.6496891 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.15897056 |
| FWER p-Value | 0.251 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | GRB7 | 20673 | 22 | 9.600 | 0.1296 | Yes | ||
| 2 | LCK | 15746 | 27 | 9.283 | 0.2559 | Yes | ||
| 3 | STAT1 | 3936 5524 | 173 | 6.741 | 0.3399 | Yes | ||
| 4 | GAB1 | 18828 | 280 | 5.825 | 0.4136 | Yes | ||
| 5 | VEGFA | 22969 | 560 | 4.131 | 0.4548 | Yes | ||
| 6 | MAPK1 | 1642 11167 | 646 | 3.755 | 0.5014 | Yes | ||
| 7 | PTEN | 5305 | 661 | 3.694 | 0.5510 | Yes | ||
| 8 | STAT3 | 5525 9906 | 681 | 3.642 | 0.5995 | Yes | ||
| 9 | STAT5A | 20664 | 824 | 3.180 | 0.6352 | Yes | ||
| 10 | RASA1 | 10174 | 929 | 2.929 | 0.6695 | Yes | ||
| 11 | NCK2 | 9448 | 1018 | 2.695 | 0.7015 | Yes | ||
| 12 | PRKCD | 21897 | 1332 | 2.074 | 0.7129 | Yes | ||
| 13 | SLC9A3R1 | 11250 | 1679 | 1.595 | 0.7160 | Yes | ||
| 14 | RAC1 | 16302 | 1922 | 1.265 | 0.7202 | Yes | ||
| 15 | GRB2 | 20149 | 2633 | 0.675 | 0.6911 | No | ||
| 16 | ITGAV | 4931 | 2923 | 0.491 | 0.6822 | No | ||
| 17 | SHB | 10493 | 2962 | 0.469 | 0.6866 | No | ||
| 18 | MYC | 22465 9435 | 3081 | 0.419 | 0.6859 | No | ||
| 19 | FYN | 3375 3395 20052 | 3206 | 0.359 | 0.6841 | No | ||
| 20 | PDGFRB | 23539 | 3809 | 0.189 | 0.6543 | No | ||
| 21 | SPHK1 | 5484 1266 1402 | 3834 | 0.183 | 0.6555 | No | ||
| 22 | ABL1 | 2693 4301 2794 | 3847 | 0.181 | 0.6573 | No | ||
| 23 | CSK | 8805 | 3853 | 0.180 | 0.6595 | No | ||
| 24 | FGR | 4723 | 4353 | 0.105 | 0.6340 | No | ||
| 25 | SNX15 | 23993 3761 | 4689 | 0.079 | 0.6170 | No | ||
| 26 | MAPK3 | 6458 11170 | 4819 | 0.071 | 0.6110 | No | ||
| 27 | SHC1 | 9813 9812 5430 | 4928 | 0.066 | 0.6061 | No | ||
| 28 | PIK3R1 | 3170 | 5108 | 0.059 | 0.5973 | No | ||
| 29 | HCK | 14787 | 5214 | 0.056 | 0.5924 | No | ||
| 30 | SOS1 | 5476 | 5642 | 0.043 | 0.5700 | No | ||
| 31 | PIK3CA | 9562 | 5729 | 0.040 | 0.5659 | No | ||
| 32 | STAT5B | 20222 | 5983 | 0.035 | 0.5527 | No | ||
| 33 | GNAO1 | 4785 3829 | 5992 | 0.034 | 0.5527 | No | ||
| 34 | PLCB2 | 5262 | 6166 | 0.031 | 0.5438 | No | ||
| 35 | PDGFB | 22199 2311 | 6490 | 0.025 | 0.5268 | No | ||
| 36 | PTPN11 | 5326 16391 9660 | 6513 | 0.024 | 0.5259 | No | ||
| 37 | PTGS2 | 5317 9655 | 6945 | 0.018 | 0.5029 | No | ||
| 38 | DOK1 | 17104 1018 1177 | 7081 | 0.015 | 0.4959 | No | ||
| 39 | KDR | 16509 | 7477 | 0.010 | 0.4747 | No | ||
| 40 | LYN | 16281 | 7605 | 0.008 | 0.4680 | No | ||
| 41 | GNAZ | 71 | 7698 | 0.007 | 0.4631 | No | ||
| 42 | YES1 | 5930 | 7788 | 0.005 | 0.4584 | No | ||
| 43 | HIF1A | 4850 | 8051 | 0.002 | 0.4443 | No | ||
| 44 | GNAI1 | 9024 | 9647 | -0.018 | 0.3586 | No | ||
| 45 | GNAI3 | 15198 | 10128 | -0.025 | 0.3330 | No | ||
| 46 | SRC | 5507 | 10226 | -0.026 | 0.3282 | No | ||
| 47 | PLCG1 | 14753 | 10828 | -0.036 | 0.2963 | No | ||
| 48 | LRP1 | 9284 | 11154 | -0.042 | 0.2793 | No | ||
| 49 | NCK1 | 9447 5152 | 11287 | -0.044 | 0.2728 | No | ||
| 50 | PTPN2 | 23414 | 11514 | -0.049 | 0.2613 | No | ||
| 51 | VAV2 | 5848 2670 | 11856 | -0.057 | 0.2437 | No | ||
| 52 | GRB10 | 4799 | 12295 | -0.070 | 0.2210 | No | ||
| 53 | ACP1 | 4317 | 12942 | -0.093 | 0.1875 | No | ||
| 54 | AKT1 | 8568 | 13376 | -0.114 | 0.1657 | No | ||
| 55 | DNM2 | 4635 3103 | 13648 | -0.133 | 0.1529 | No | ||
| 56 | RHOA | 8624 4409 4410 | 13936 | -0.158 | 0.1396 | No | ||
| 57 | BCAR1 | 18741 | 14108 | -0.174 | 0.1327 | No | ||
| 58 | SLC9A3R2 | 23089 1544 1514 | 14485 | -0.223 | 0.1155 | No | ||
| 59 | CBL | 19154 | 15001 | -0.315 | 0.0920 | No | ||
| 60 | PTPRJ | 9664 | 15136 | -0.360 | 0.0897 | No | ||
| 61 | BLK | 3205 21791 | 15569 | -0.514 | 0.0734 | No | ||
| 62 | ITGB3 | 20631 | 16124 | -0.865 | 0.0554 | No | ||
| 63 | HRAS | 4868 | 16218 | -0.935 | 0.0631 | No | ||
| 64 | PTPN1 | 5325 | 18317 | -4.853 | 0.0161 | No |

